Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Natsui, Yu*; Nefissi, R.*; Miyata, Kana*; Oda, Atsushi*; Hase, Yoshihiro; Nakagawa, Mayu; Mizoguchi, Tsuyoshi*
no journal, ,
EARLY FLOWERING 3 (ELF3) plays key roles in the control of light response, organ elongation, flowering time and circadian rhythms in Arabidopsis. ELF3 does not show significant homology to any other proteins in the public databases. Therefore biochemical function of the ELF3 has not been well understood. To understand molecular mechanisms underlying the a variety of phenotypes in the elf3 mutants, suppressors of elf3-1 were screened for, using the elf3-1 seeds mutagenized with heavy ion beams. Here we demonstrate that in one of the suppressor lines, S#20, early flowering, small cotyledon and pale green phenotypes of elf3-1 were suppressed, whereas long hypocotyl phenotype was enhanced. The recessive suppressor mutation was named suppressor of elf3 20 (sel20) and mapped to the upper side of Ch1. We have found 21-bp deletion in the FHA/CRY2 gene in the sel20. We will discuss possible roles of CRY2 in the multiple functions of ELF3.
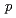 -chlorophenoxyisobutyric acid, resistant mutants
-chlorophenoxyisobutyric acid, resistant mutantsOno, Yutaka
no journal, ,
We have shown that the putative antiauxin 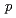 -chlorophenoxyisobutyric acid impairs auxin-induced gene expression and AUX/IAA protein degradation in Arabidopsis root. We have also found that root growth of the Arabidopsis auxin signaling mutants is less inhibited by PCIB than that of wild type or the auxin transport mutants. The effort to screen auxin signaling mutants whose root growth is not inhibited by PCIB led identification of the novel mutants,
-chlorophenoxyisobutyric acid impairs auxin-induced gene expression and AUX/IAA protein degradation in Arabidopsis root. We have also found that root growth of the Arabidopsis auxin signaling mutants is less inhibited by PCIB than that of wild type or the auxin transport mutants. The effort to screen auxin signaling mutants whose root growth is not inhibited by PCIB led identification of the novel mutants, 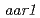 and
and 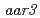 , as well as known auxin loci,
, as well as known auxin loci, 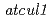 and
and 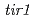 . Both
. Both 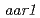 and
and 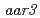 are also resistant to 2,4-D. The protein encoded by
are also resistant to 2,4-D. The protein encoded by 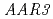 shares homology with the DCN-1 protein (RUB E3 ligase).
shares homology with the DCN-1 protein (RUB E3 ligase). 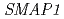 , the gene responsible for
, the gene responsible for 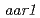 , was functionally unknown. However, our experiments suggested that the SMAP1 protein is also involved in RUB modification. These results indicate that PCIB could facilitate the identification of factors involved in auxin-related signaling.
, was functionally unknown. However, our experiments suggested that the SMAP1 protein is also involved in RUB modification. These results indicate that PCIB could facilitate the identification of factors involved in auxin-related signaling.
Kitamura, Satoshi; Toge, Takayuki*; Matsuda, Fumio*; Sakakibara, Keiko*; Saito, Kazuki*; Narumi, Issei
no journal, ,
Flavonoids are widely distributed in the plant kingdom, and their functions as antimicrobial agents and UV-B protectants are essential for the normal growth of plants under stressful environments. Flavonoid biosynthesis is thought to proceed in the cytosolic surface of the endoplasmic reticulum, whereas many endproducts of flavonoid are accumulated in vacuoles. In the model plant Arabidopsis, intracellular flavonoid transport mechanisms are investigated for proanthocyanidin pathway, one of the flavonoid subclass being synthesized and accumulated specifically in seed coat. Some factors presumably involved in proanthocyanidin transport/accumulation, such as membrane-bound proanthocyanidin transporter TT12 and putative proanthocyanidin ligandin TT19, have so far been isolated. We describe here the results on metabolic analysis of immature seed extracts from these flavonoid mutants.
Kawachi, Naoki; Suzui, Nobuo; Ishii, Satomi; Ito, Sayuri; Watanabe, Shigeki; Ishioka, Noriko; Takahashi, Tadayuki*; Nakano, Takashi*; Fujimaki, Shu
no journal, ,
no abstracts in English
Ishii, Satomi; Suzui, Nobuo; Ito, Sayuri; Kawachi, Naoki; Ishioka, Noriko; Otake, Norikuni*; Oyama, Takuji*; Fujimaki, Shu
no journal, ,
no abstracts in English
Nakasone, Akari; Kawai, Maki*; Narumi, Issei; Uchimiya, Hirofumi*; Ono, Yutaka
no journal, ,
The 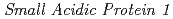 (
(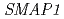 ) gene is recently identified as the factor that mediates the synthetic auxin 2,4-D response in
) gene is recently identified as the factor that mediates the synthetic auxin 2,4-D response in 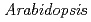 root. SMAP1 has the conserved phenylalanine and asparagic acid (F/D) domain, which is highly conserved in both plants and animals. Introduction of
root. SMAP1 has the conserved phenylalanine and asparagic acid (F/D) domain, which is highly conserved in both plants and animals. Introduction of 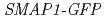 to
to 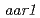 recovered 2,4-D sensitivity of
recovered 2,4-D sensitivity of 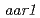 while that of
while that of 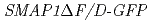 did not. Using anti-GFP-MicroBeads, the proteins that form complexes with SMAP1-GFP but not with SMAP1
did not. Using anti-GFP-MicroBeads, the proteins that form complexes with SMAP1-GFP but not with SMAP1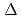 F/D-GFP were pulled down from the
F/D-GFP were pulled down from the  extract and identified by MS analysis. The identified peptide sequences indicated that SMAP1-GFP binds to COP9 Signalosome (CSN). The results implied that SMAP1 regulates 2,4-D response by interacting to CSN through F/D region.
extract and identified by MS analysis. The identified peptide sequences indicated that SMAP1-GFP binds to COP9 Signalosome (CSN). The results implied that SMAP1 regulates 2,4-D response by interacting to CSN through F/D region.
Sakamoto, Ayako; Nakagawa, Mayu; Narumi, Issei
no journal, ,
no abstracts in English
Takano, Nao*; Takahashi, Yuko*; Yamamoto, Mitsuru*; Teranishi, Mika*; Yokozawa, Daisuke*; Hase, Yoshihiro; Sakamoto, Ayako; Tanaka, Atsushi; Hidema, Jun*
no journal, ,
no abstracts in English